Research Article - (2022) Volume 11, Issue 3
Background: Rhesus monkeys (Macaca mulatta) are extensively used in the field of medical and psychological research as a valuable model organism. To explore the breeding potential and exchange of germplasm resources for rhesus monkeys, genetic diversity derived from two different captive populations was comparatively evaluated based on polymorphic microsatellite markers.
Methods: Fifteen polymorphic microsatellite markers were used to analyze the genetic diversity parameters. Micro-Checker was used to check for genotyping errors due to null alleles, allelic dropout, and stuttering. Hardy– Weinberg equilibrium (HWE) test were analyzed using Genepop on the web. The best K value was calculated by Structure Harvester, CLUMPP_Windows and distruct were used to visualize the individual coefficients of membership in the subpopulation. PcoA and Molecular variance analysis (AMOVA) analysis were used by GenAlex.
Results: A total of 155 alleles were identified, with the number of alleles per locus ranging from 7 to 15, giving an average number of 10.3 alleles per locus. The mean Number of effective alleles (Ne), observed Heterozygosity (Ho), expected Heterozygosity (He), and the Polymorphism Information Content (PIC) were 5.602, 0.7297, 0.8016, and 0.7716, respectively. The populations Hengshu (Hereafter HS) and Xijie (hereafter XJ) shared partial common alleles; however, the other left alleles in XJ were not detected. Structure analysis indicated that two populations belong to three genetic lineages. AMOVA showed that the genetic variance was 91% within individuals, while it was 9% among populations, respectively.
Conclusion: Higher genetic diversity level of captive population in HS than that in XJ, population structure analysis showed two rhesus monkey populations had no significant genetic differentiation. These results can provide a beneficial reference for the exchange of germplasm resources and genetic management.
Macaca mulatta • Microsatellite • Genetic diversity • Population structure
Rhesus monkeys (Macaca mulatta) are classified in the family Cercopithecidae (the Old World monkeys) [1], they were considered to be one of the most evolutionarily successful and intensively studied nonhuman primate species. The phylogenetic tree indicated that they shared a common ancestor about 25 million years ago [2]. They live in groups consisting of a single adult male along with several females and their offspring; males leave the troop at maturity, whereas females tend to stay in the troops in which they were born, which belong to the polygyny marriage and male-biased dispersal. Rhesus monkeys occupy a great diversity of altitudes and a great variety of habitats, from grasslands to arid and forested areas, but also close to human settlements. In China, there are a total of six rhesus monkey subspecies, which are all classified and listed in the second class of the Chinese key protected wildlife. Wild rhesus monkeys in the northernmost and southernmost are mainly distributed in Taihang Mountain and Hainan island of China, respectively.
Due to their similarities to humans in anatomical and physiological traits, behaviors, and genetic materials, rhesus monkeys are allowed professional artificial breeding as model organisms. They are extensively used in the field of medical and psychological research as valuable experimental animals. Experiment monkeys are designed and serve as an animal model of human diseases, which are mainly embodied in the diabetes model, Parkinson’s and Alzheimer's diseases, obesity model [3], and AIDS model etc. Moreover, other research related to rhesus monkeys is also involved in cloning [4], investigation of infectious diseases and vaccine development [5], stem cell research, organ transplants, and comparative genomics [6].
Microsatellites, also known as Short Tandem Repeats (STR), are highly polymorphic DNA fragments composed of repetitive stretches of short sequences of 1-6 base pairs of DNA, which serve as valuable molecular markers to track inheritance within families. The polymorphisms originated from the number of the tandem repeats of a specific microsatellite at a specific locus, which is distinguished from the microsatellites by the size of core motifs (few nucleotides vs. several tens). By comparison, SSR marker has many advantages including high abundance, random distribution in the entire genome, high information content, codominant inheritance, and reproducibility, which can reconstruct the genetic relationship among individuals, identifies specific species, and the blood relationship between individuals, etc. Therefore, they were preferred as the most popular and effective genetic markers in population census, estimating genetic diversity [7], diffusion model, population management, and paternity test. Nowadays, the applications of fluorescent microsatellite markers further contribute to reflecting the microsatellite variance of allele size accurately on the chromosomal level.
Genetic diversity is the basis for the survival and evolution of species. The abundance of genetic diversity of a species is mainly affected by three factors: Small population, bottleneck, and gene flow. The rich genetic diversity and genetic variation within a species is conducive to improving the ability of species to adapt to environmental changes. Decreased genetic diversity will lead to reduced population adaptability and viability, and even increase the risk of extinction. As an important component for both the short and long-term persistence of the experimental rhesus monkey populations, the assessment of genetic diversity is a problem worthy to be explored. Furthermore, insights into the genetic diversity and structure of captive rhesus monkeys would provide a theoretical basis for conservation, breeding, and sustainable development.
Sample collection and DNA extraction
A total of 104 blood samples for routine examination were collected from two captive Rhesus monkey populations (HS=69 and XJ=35). The extraction of genomic DNA was conducted using a TianGen DNA extraction kit (Beijing), and the final genomic DNA was used for electrophoresis on 1% agarose gel stained with Golden View (Shanghai) to assess the DNA quality.
Microsatellite loci amplification and genotyping
In this study, fifteen novel tetranucleotide perfect microsatellite loci with high polymorphisms (PIC>0.5) were selected for identifying the variation of allele fragments [8]. Based on a suitable annealing temperature, all the forward primers were labeled with different fluorescent dyes (FAM or HEX) and then used for amplification and genotyping in 104 Rhesus monkeys. Detailed characteristics and information of 15 polymorphic microsatellite loci were shown in Table 1.
| Chromosome ID | Loci | Primer sequences (5'- 3') | Core motif | Fluorescent dyes | Ta (℃) | Product (bp) | Genbank accession No. |
|---|---|---|---|---|---|---|---|
| chr1 | C1 033 | F:AACTTCACCAAGATGCACTG | (TATC)12 | Fam | 57.1 | 248bp | MF045896 |
| R:TGGTATTGTGGTTATGTAGGG | |||||||
| chr2 | C2 038 | F:AGTGAATGGAGAACAGAAATGC | (ATCT)14 | Hex | 60 | 229bp | MF045897 |
| R:CATGATTCAGCAACTGCCTT | |||||||
| chr3 | C3 001 | F:TGCCTGTTCACTCTGATGGT | (TCTA)14 | Fam | 62.5 | 281bp | MF045898 |
| R:TGGAGGGGTGATACTGGTG | |||||||
| chr4 | C4 001 | F:TCCAGTTACTTTCCCCAGAGC | (CATT)16 | Hex | 62.5 | 171bp | MF045899 |
| R:AAGACAGCGGCATAGAGGC | |||||||
| chr5 | C5 009 | F:ACACCCTTATCACCCATCAG | (AAGG)12 | Fam | 62.7 | 213bp | MF045900 |
| R:TCCCCTCCCATTTCCCCTTC | |||||||
| chr6 | C6 001 | F:TGAATCAAGGATGGACGGA | (GGAT)16 | Hex | 54 | 244bp | MF045901 |
| R:GGGGACTTAGAGCCCACAAT | |||||||
| chr8 | C8 013 | F:GCAGAAAGCAGACAGCCTATT | (CATC)10 | Hex | 61 | 213bp | MF045902 |
| R:GGATGCGTGGATATGTGG | |||||||
| chr9 | C9 002 | F:CCCTGGTTCTTGTCCTAAATG | (AGAT)14 | Fam | 60 | 220bp | MF045903 |
| R:TCCCTGGAGGAATCTGTGG | |||||||
| chr10 | C10 016 | F:GGAGACTGAGGCACAACAATC | (AAAT)11 | Hex | 62.5 | 215bp | MF045904 |
| R:GCAAGTGAAATGCTAACCAAC | |||||||
| chr11 | C11 019 | F:AAAGTGTAGAGGGTCAAGATGC | (AAAG)19 | Fam | 62.7 | 256bp | MF045905 |
| R:GAGGTTGAAAAGGGTTTGTTTG | |||||||
| chr13 | C13 016 | F:GGGAGCAAGCAAACAACAT | (ATCT)15 | Fam | 66 | 211bp | MF045908 |
| R:CAGCAAAGGATAGACAGGTGAT | |||||||
| chr14 | C14 011 | F:CCTTTGTTCACTGAGCAGCA | (TCCT)14 | Fam | 66 | 258bp | MF045909 |
| R:GGCAGGAGAATCACTTGGAC | |||||||
| chr15 | C15 019 | F:GAGTAGAGCAAGCCTTGGAAC | (TATC)14 | Fam | 57.9 | 215bp | MF045910 |
| R:AGGAGAATCACCTAAACCCAG | |||||||
| chr17 | C17 010 | F:AACATCAGTTATCAGGGAAGGA | (ATTT)19 | Hex | 62.5 | 286bp | MF045912 |
| R:GAGGCTGGGTTAGGAGGAT | |||||||
| chr18 | C18 021 | F:CTCCAGTGATCCTCCTGATTC | (TTAG)16 | Hex | 65 | 203bp | MF045913 |
| R:ATGAGCCAAGTGAGACTCCAT |
Table 1. Chromosome ID, Primer sequences, core motif, fluorescent dyes, annealing temperatures, product length, accession number of 15 microsatellites loci.
PCR was carried out in a 25-uL reaction mixture comprising 1.0 μL of genomic DNA, 0.25 μL of MgCl2 (TaKaRa, Dalian), 1.0 μL of each dNTP (Vazyme, Nanjing), 1 μL of each primer, 3.0 μL of PCR buffer (Vazyme), and 0.25 μL of Taq DNA polymerase (Vazyme), with deionized water used to make the sample volume up to 25 μL. The reactions were performed using an initial denaturation at 95℃ for 4 min, followed by 30 cycles at 94℃ for 30 s, specific annealing for 30 s, primer extension at 72℃ for 35 s, and a final extension at 72℃ for 10 min. Subsequently, the reaction products were kept at 4℃. After amplification, 3.0 μL of each PCR product was used for electrophoresis on 1.5% agarose gel stained with GoldenView (Biomed) to detect the PCR quality. ABI Prism 377 (Applied Bio systems) was used for genotyping using the GeneScan Tamara 350 internal size standard (ABI) for PCR products.
Genetic diversity and population structure analysis
The peak shape was accurately analyzed, and the allele size was manually corrected. Micro-Checker 2.2.3 was used to check for genotyping errors due to null alleles, allelic dropout, and stuttering. The number of alleles (N), observed Heterozygosity (Ho), expected Heterozygosity (He), and the Polymorphism Information Content (PIC) were calculated using Cervus 3.0. Null allele frequency F (null) and P-value for Hardy-Weinberg Equilibrium (HWE) test were analyzed using Genepop on the web. In addition, Structure 2.3.4 was used to infer the potential populations (K), the Length of the Burnin Period, and the Number of MCMC (Monte Carlo Markov chain cycles) Reps after Burnin were set as 100,000 and 1,000,000 using the admixture model of independent allele frequencies, respectively. And the variation range of the K value was set from 2 to 7. For each K value, it ran 10 times independently. The best K value was calculated by Structure Harvester, CLUMPP_Windows.1.1.2, and distruct 1.1 were used to visualize the individual coefficients of membership in the subpopulation. PcoA and Molecular variance analysis (AMOVA) analysis were used by GenAlex 6.503.
Genetic diversity analysis in the whole population
A total of novel fifteen microsatellite markers were used to analyze the genetic diversity of the captive Rhesus monkeys population, and the characteristics of microsatellite loci were summarized in Table 2. The number of observed alleles (Na) is one of the most important indicators of gene differentiation, which is directly related to population, type, and geographic location. The analysis results of 15 microsatellite loci showed 155 alleles were identified in the 104 individuals, with the number of alleles each locus ranging from 7 (C4 001 and C17 010) to 17 (C13 016), giving an average number of 10.3 alleles per locus. The number of effective alleles in each locus ranged from 3.401 to 10.989, the mean effective number of alleles was 5.602. Heterozygosity present in populations reflects the genetic variation of the population at different loci, which can be used as an optimal parameter to evaluate the genetic diversity. The observed ranged from 0.409 to 0.891 and expected heterozygosity ranged from 0.706 to 0.909, respectively. Mean Ho and He were 0.7297 and 0.8016, respectively. When the heterozygosity is between 0.5 and 0.8, it can be considered that the genetic diversity of the population is high. The PIC ranged from 0.663 to 0.897 with an average value of 0.771. Moreover, nine out of fifteen loci presented null alleles frequencies F(Null), ranging from - 0.0289 to 0.3801 (Table 3).
| Chr ID | Primer | Size (bp) | N | Ne | Ho | He | PIC | P value | F(Null) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | C1 033 | 223-255 | 8 | 3.401 | 0.591 | 0.706 | 0.663 | 0.0464 | 0.0903 |
| 2 | C2 038 | 220-248 | 8 | 4.201 | 0.755 | 0.762 | 0.725 | 0.1627 | -0.0005 |
| 3 | C3 001 | 254-282 | 8 | 4.807 | 0.809 | 0.792 | 0.759 | 0.6071 | -0.0169 |
| 4 | C4 001 | 129-153 | 7 | 4.347 | 0.755 | 0.77 | 0.727 | 0.3922 | 0.0057 |
| 5 | C5 009 | 191-231 | 10 | 5.181 | 0.691 | 0.807 | 0.776 | 0.0198 | 0.0808 |
| 6 | C6 001 | 190-246 | 13 | 4.672 | 0.655 | 0.786 | 0.758 | 0 | 0.0963 |
| 8 | C8 013 | 196-228 | 9 | 4.405 | 0.809 | 0.773 | 0.734 | 0.5527 | -0.0278 |
| 9 | C9 002 | 196-256 | 15 | 8 | 0.891 | 0.875 | 0.857 | 0.7632 | -0.0121 |
| 10 | C10 016 | 188-220 | 9 | 3.875 | 0.764 | 0.742 | 0.714 | 0.723 | -0.0127 |
| 11 | C11 019 | 225-285 | 15 | 8.403 | 0.745 | 0.881 | 0.865 | 0.0051 | 0.0811 |
| 13 | C13 016 | 187-261 | 17 | 10.989 | 0.409 | 0.909 | 0.897 | 0 | 0.3801 |
| 14 | C14 011 | 233-269 | 10 | 7.194 | 0.827 | 0.861 | 0.841 | 0.1998 | 0.017 |
| 15 | C15 019 | 200-236 | 10 | 5.917 | 0.745 | 0.831 | 0.806 | 0.1439 | 0.0515 |
| 17 | C17 010 | 232-256 | 7 | 4.854 | 0.836 | 0.794 | 0.759 | 0 | -0.0289 |
| 18 | C18 021 | 157-189 | 9 | 3.787 | 0.664 | 0.736 | 0.694 | 0.12 | 0.0525 |
| Mean | - | - | 10.3 | 5.602 | 0.7297 | 0.8016 | 0.7716 | 0.745 | 0.718 |
Table 2. Total genetic diversity parameters of 15 microsatellite markers.
| Pop | Locus | Na | Ne | I | Ho | He | uHe | F | P-value |
|---|---|---|---|---|---|---|---|---|---|
| HS | C1033 | 8 | 3.409 | 1.473 | 0.646 | 0.707 | 0.712 | 0.086 | 0.1076 |
| (N=65) | C2038 | 7 | 3.815 | 1.534 | 0.815 | 0.738 | 0.744 | -0.105 | 0.6449 |
| C3001 | 8 | 3.967 | 1.609 | 0.754 | 0.748 | 0.754 | -0.008 | 0.6857 | |
| C4001 | 7 | 4.191 | 1.526 | 0.769 | 0.761 | 0.767 | -0.01 | 0.9942 | |
| C5009 | 10 | 4.904 | 1.794 | 0.692 | 0.796 | 0.802 | 0.13 | 0.0102 | |
| C6001 | 12 | 4.763 | 1.842 | 0.708 | 0.79 | 0.796 | 0.104 | 0.009 | |
| C8013 | 9 | 4.499 | 1.67 | 0.8 | 0.778 | 0.784 | -0.029 | 0.286 | |
| C9002 | 15 | 8.086 | 2.259 | 0.908 | 0.876 | 0.883 | -0.036 | 0.2664 | |
| C10016 | 9 | 3.641 | 1.672 | 0.708 | 0.725 | 0.731 | 0.024 | 0.1547 | |
| C11019 | 15 | 8.527 | 2.304 | 0.8 | 0.883 | 0.89 | 0.094 | 0.2179 | |
| C13016 | 17 | 11.45 | 2.601 | 0.369 | 0.913 | 0.92 | 0.595 | 0 | |
| C14011 | 10 | 7.119 | 2.063 | 0.831 | 0.86 | 0.866 | 0.033 | 0.2499 | |
| C15019 | 10 | 6.006 | 1.96 | 0.785 | 0.833 | 0.84 | 0.059 | 0.3239 | |
| C17010 | 7 | 4.782 | 1.656 | 0.846 | 0.791 | 0.797 | -0.07 | 0.0013 | |
| C18021 | 7 | 3.539 | 1.474 | 0.677 | 0.717 | 0.723 | 0.056 | 0.1916 | |
| Mean ± SE | 10.067 ± 0.842 | 5.513 ± 0.598 | 1.829 ± 0.088 | 0.741 ± 0.032 | 0.794 ± 0.017 | 0.801 ± 0.017 | 0.062 ± 0.042 | - | |
| XJ | C1033 | 7 | 2.931 | 1.372 | 0.513 | 0.659 | 0.667 | 0.222 | 0.0849 |
| (N=39) | C2038 | 7 | 4.237 | 1.6 | 0.641 | 0.764 | 0.774 | 0.161 | 0.2125 |
| C3001 | 8 | 5.633 | 1.876 | 0.872 | 0.822 | 0.833 | -0.06 | 0.836 | |
| C4001 | 6 | 4.156 | 1.55 | 0.769 | 0.759 | 0.769 | -0.013 | 0.0188 | |
| C5009 | 7 | 5.263 | 1.774 | 0.667 | 0.81 | 0.821 | 0.177 | 0.0537 | |
| C6001 | 12 | 4.768 | 1.913 | 0.641 | 0.79 | 0.801 | 0.189 | 0.0038 | |
| C8013 | 6 | 4.111 | 1.521 | 0.846 | 0.757 | 0.767 | -0.118 | 0.8867 | |
| C9002 | 9 | 5.805 | 1.9 | 0.846 | 0.828 | 0.838 | -0.022 | 0.5575 | |
| C10016 | 7 | 3.583 | 1.546 | 0.821 | 0.721 | 0.73 | -0.138 | 0.8702 | |
| C11019 | 10 | 7.493 | 2.101 | 0.641 | 0.867 | 0.878 | 0.26 | 0.0017 | |
| C13016 | 12 | 8.199 | 2.239 | 0.513 | 0.878 | 0.889 | 0.416 | 0 | |
| C14011 | 9 | 6.17 | 1.97 | 0.795 | 0.838 | 0.849 | 0.051 | 0.3967 | |
| C15019 | 9 | 5.13 | 1.863 | 0.718 | 0.805 | 0.816 | 0.108 | 0.4845 | |
| C17010 | 6 | 4.651 | 1.64 | 0.821 | 0.785 | 0.795 | -0.045 | 0.0134 | |
| C18021 | 7 | 3.935 | 1.547 | 0.667 | 0.746 | 0.756 | 0.106 | 0.0771 | |
| Mean ± SE | 8.133 ± 0.515 | 5.071 ± 0.368 | 1.761 ± 0.063 | 0.718 ± 0.030 | 0.789 ± 0.014 | 0.799 ± 0.014 | 0.086 ± 0.040 | - |
Table 3. Genetic diversity indices of 15 microsatellite loci in two captive populations.
Comparison of genetic diversity between HS and XJ population
The number of alleles at each locus (Na), the number of effective alleles (Ne), Shannon index (I), observed Heterozygosity (Ho), expected Heterozygosity (He), P-value for Hardy-Weinberg equilibrium for HS (N=65), and XJ (N=39) were summarized in Table 4. The number of alleles on each locus in HS was greater than or equal to that in XJ; in particular, locus C3 001, C4 001, and C18 021 shared the same alleles. The Ne ranged from 3.409 to 11.45 in HS, and ranged from 2.931 to 8.199 in XJ, respectively. The mean Ho of the HS group (Ho=0.741)was higher than that of the XJ group (Ho=0.718). Besides, the mean (He ~ 0.745) and PIC (~ 0.761) were also lower in the XJ group than in the HS group (He=0.801, PIC=0.767). To sum up, we speculate the two captive Rhesus monkeys possess a relatively high level of genetic diversity. HWE analysis showed that 4 loci (C5 009, C6 001, C13 016, and C17 010) in HS population and 5 loci (C4 001, C6 001, C11 019, C13 016, and C17 010) in XJ population, and the remaining loci were in accordance with HWE.
| Locus | Fis | Fit | Fst | Nm |
|---|---|---|---|---|
| C1033 | 0.151 | 0.154 | 0.004 | 68.677 |
| C2038 | 0.03 | 0.035 | 0.005 | 46.054 |
| C3001 | -0.035 | -0.024 | 0.011 | 22.117 |
| C4001 | -0.012 | -0.007 | 0.004 | 58.768 |
| C5009 | 0.154 | 0.156 | 0.003 | 90.209 |
| C6001 | 0.147 | 0.15 | 0.004 | 65.963 |
| C8013 | -0.073 | -0.07 | 0.002 | 110.719 |
| C9002 | -0.029 | -0.02 | 0.009 | 28.358 |
| C10016 | -0.057 | -0.052 | 0.005 | 54.234 |
| C11019 | 0.176 | 0.178 | 0.002 | 122.271 |
| C13016 | 0.507 | 0.511 | 0.007 | 34.355 |
| C14011 | 0.042 | 0.046 | 0.004 | 60.721 |
| C15019 | 0.083 | 0.085 | 0.002 | 107.796 |
| C17010 | -0.058 | -0.054 | 0.003 | 75.092 |
| C18021 | 0.082 | 0.083 | 0.001 | 216.504 |
| Mean ± SE | 0.074 ± 0.038 | 0.078 ± 0.038 | 0.004 ± 0.001 | 77.456 ± 12.580 |
Table 4. F-statistics analysis and Nm index of 15 microsatellite loci.
Analysis of Molecular Variance(AMOVA)is a method to detect population differentiation utilizing molecular markers. This procedure was initially implemented for DNA haplotypes but applies to any marker system. The implementation of AMOVA requires two very basic components: (1) A distance matrix derived from the data and (2) a separate table used to partition the data into different stratifications. AMOVA showed that the genetic variance was 91% within individuals, while it was 9% among populations, respectively (Figure 1).
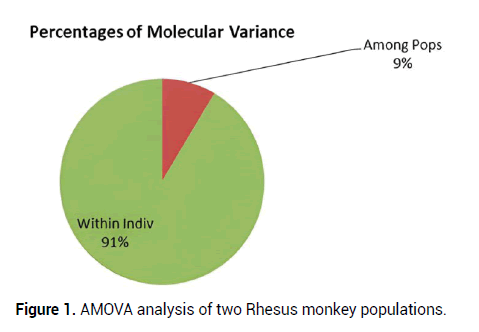
Figure 1: AMOVA analysis of two Rhesus monkey populations.
Population structure analysis
Animals in captivity are also subject to similar evolutionary forces that act on natural populations facilitating the generation of population genetic structure. Population genetic structure essentially describes the total genetic diversity and its distribution within and among a set of populations. Given that the introduction of wild Rhesus monkeys individuals, we wonder whether there existed the peculiar allele inherited and retained in some wild individuals. To address this, the Bayesian method of Structure 2.3.4 was carried out to analyze the genetic structure of the captive population. We speculated that the captive population received frequent gene flow and introgression due to artificial selection. Given the artificial interference and the introduction of wild individuals, Structure analysis showed when K=3, the Delta K estimator exhibited an obvious apex Delta (K=0.689839) (Figure 2). The assignment results show K=3, three colors represent three different genetic clusters. The three colors red, blue, and yellow, represent three ancestral blood lineages and are distributed in all the samples. Each line represents one individual, and the proportion of population assignment of each individual is relative to the given genetic cluster, which is represented by the length of each line (Figure 3). In the ideal situation, each column represented one individual and the colors represented the probability membership coefficient of that individual for the genetic cluster, however, no obvious genetic difference was found among the captive individuals.
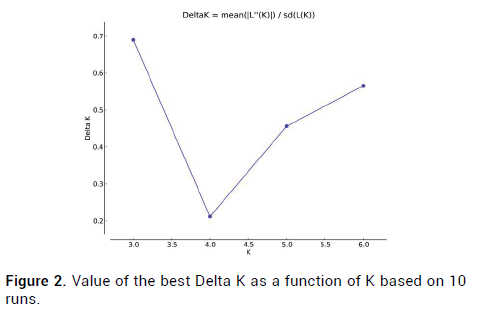
Figure 2: Value of the best Delta K as a function of K based on 10 runs.
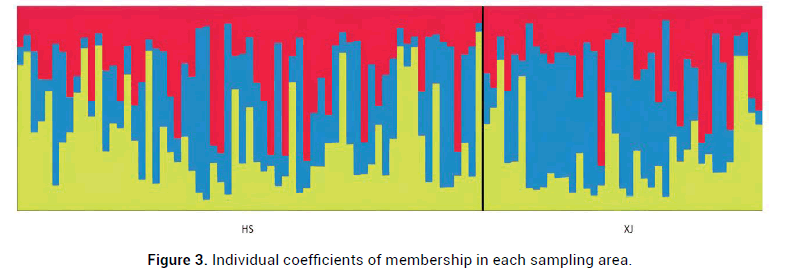
Figure 3: Individual coefficients of membership in each sampling area.
Principal coordinate analysis was used to establish a two-dimensional location map of two captive rhesus monkeys populations, which can visualize the difference or similarity of data. The results showed the first and second principal coordinates explained 6.77% (HS, Pop1) and 6.48% (XJ, Pop2) of the total variation, respectively. PCoA analysis did not separate the populations (Figure 4).
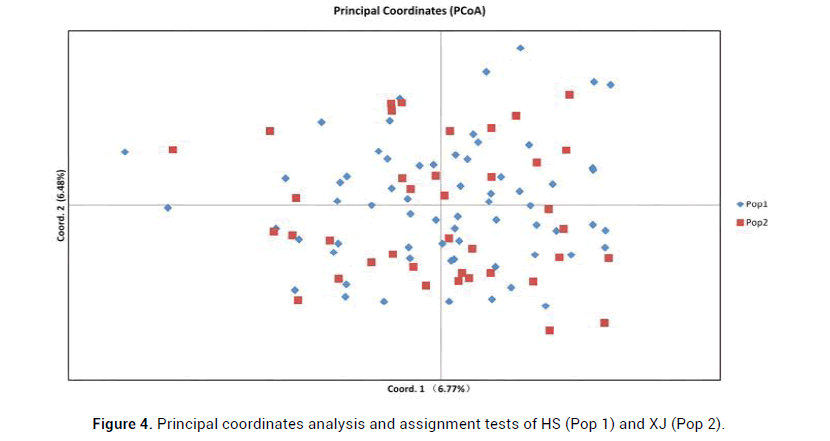
Figure 4: Principal coordinates analysis and assignment tests of HS (Pop 1) and XJ (Pop 2).
F-statistics analysis
Understanding the extent of genetic differentiation among captive populations provides insights into industry practices and the domestication process. F-statistics showed the mean Fis and Fit were 0.074 ± 0.038 and 0.078 ± 0.038, respectively, which indicated the inbreeding, is not obvious. Wright (1965) proposed that the genetic differentiation coefficient Fst<0.05 is low differentiation, 0.05 ≤ Fst ≤ 0.15 is moderate differentiation, Fst>0.15 is highly differentiated, and Fst>0.25 is extremely differentiated [9]. In this study, the Fst was 0.001 to 0.011, less than 0.05, which was a low degree of differentiation. Gene flow can play a homogenizing role in the population and effectively resist genetic differentiation caused by selection and genetic drift. Theoretically, when Nm<1, differentiation may occur among populations due to genetic drift. If Nm>1, gene exchange between populations can prevent population differentiation caused by genetic drift. In this study, gene flow between all populations ranged from 22.117 to 216.504, indicating that frequent gene exchange existed in captive monkeys under artificial selection, which could prevent population structure due to genetic drift between populations.
Selection advantage of microsatellites
Regular conservation genetics techniques for protecting endangered species mainly focused on analyzing mitochondrial DNA or microsatellites. However, studies in recent years have shown that the use of microsatellite molecular markers will more truly reflect the current status and geographic distribution patterns of endangered species' genetic diversity. This is because mitochondrial DNA is matrilineal. Genetic molecular markers can only reflect the phylogenetic history of the maternal line; moreover, among many individuals of endangered animals, mitochondrial DNA control regions rarely undergo base substitution mutations. It is more suitable for detecting genetic variation caused by long-term events, and sufficient genetic information cannot be obtained for the variation caused by recent events. Microsatellites are located in the non-coding regions of nuclear genes and are inherited by parents. They usually have more genetic information than mitochondrial DNA, and can better detect genetic mutations caused by recent events. Therefore, we used microsatellite markers to detect the genetic diversity and genetic structure of the captive rhesus monkeys to provide a scientific basis for the protection and management. We also agree that the death report of microsatellites is an exaggeration in the 21st century, which still has an important place in the genomic age as they remain effective and cost-efficient markers [10].
Genetic quality evaluation of captive populations
In terms of sample types, genetic variation is assessed mainly by collecting tissues, blood, feathers, hair, or bones. Blood samples of experiment monkeys were collected during the physical examination in this study, which facilitated the accuracy of genotyping and interpretation. When using microsatellite molecular markers to detect the genetic diversity of captive animals, the sample size of the population is required to be at least 25, and our study meets the number of samples. Concrete genetic parameters, i.e., heterozygosity present in populations can reflect the genetic variation at different loci. Average heterozygosity appeared to be an optimal measure of diversity that encompassed most of this variation, and the heterozygosity deficiencies could lead to the presence of population substructure, inbreeding, and null alleles. The characteristics of the experiment population were small scale, redistribution, and unnatural selection, leading to population degradation and loss of genetic diversity. Further, some specific and valuable gene resources eventually played themselves out. In this study, captive population HS showed higher genetic diversity than XJ population, which may be attributed to different provenances, founder effect, and differential management strategies.
Population genetic structure
Population structure analysis helps to understand the process of evolution and can identify the subgroups by studying the association between genotype and allele frequency. In the assignment tests, these experiment individuals were assigned to three blood lineages. Relatively balanced blood lineages proportion suggested no obvious genetic structural attribution by the artificial selection and introducing wild individuals. Furthermore, the relatively higher heterozygosity in the HS and the XJ indicated better breeding potential in the experiment population. In some loci, the HS and XJ shared the common alleles, however, the HS occurred to be more alleles in partial loci than XJ, which may be attributed to partial breeding individuals did not participate in the productive performance or larger number in the HS group than the XJ. More loci in the XJ group with a significant deviation from the HWE also suggested the necessity of a balanced mating assignment.
Compared with large populations, a small population may be more vulnerable to extinction; especially synergistic interactions in their dynamics can lead to an extinction vortex. Appropriate and reasonable domesticating or breeding in zoos, aquariums, or breeding bases of endangered species should all be furnished with genetic management concepts as follows: firstly, the evolutionary potential should be evaluated using inbreeding degree and genetic variation; secondly, the paternity test and pedigree construction should be made clear; thirdly, the genetic match should be performed, and breeding plans should be made based on the pedigree records. Managing experiment populations is costly (i.e., shipping animals), time-consuming (processing pedigree), and risky (stress on animals or disease transmission), however, the effect and experience are worth affirming fully.
Therefore, it was necessary to improve the management and breeding strategies according to the genetic pedigree, to facilitate more fertile individuals have the opportunity to get involved in reproduction. Higher genetic diversity level of captive rhesus monkey populations was indicated in HS than that in XJ. Moreover, no significant genetic differentiation was found in population’s structure. This research can provide a beneficial reference for the exchange of germplasm resources and genetic management.
The authors acknowledge Zhengwu Wang and Xu Luo of Yibin HengShu Animal Models Resource Industry Technology Academy for providing the samples. This study was supported by the science and technology planning projects of the Educational Commission of Jiangxi Province (GJJ180225) and the National Natural Science Foundation of China (31960118).
The authors declare that they have no conflict of interest. Samples used in this project were collected during the physical examinations of laboratory SPF rhesus monkeys. Sampling permission was under the Sichuan University Institutional Animal Care and Use Committee, and sampling strategies strictly followed the animal ethics regulations of Sichuan University.
[Cross Ref] [Google Scholar] [PubMed]
[Cross Ref] [Google Scholar] [PubMed]
[Cross Ref] [Google Scholar] [PubMed]
[Cross Ref] [Google Scholar] [PubMed]
[Cross Ref] [Google Scholar] [PubMed]
[Cross Ref] [Google Scholar] [PubMed]
[Cross Ref] [Google Scholar] [PubMed]
[Cross Ref] [Google Scholar] [PubMed]
Citation: Du C, et al. Genetic Diversity and Population Structure in Two Captive Rhesus Monkey (Macaca mulatta) Populations as Revealed by Microsatellite Markers. J Biol Today's World, 2022, 11(3), 001-006
Received: 18-Apr-2022, Manuscript No. JBTW-22-61086; Editor assigned: 21-Apr-2022, Pre QC No. JBTW-22-61086(PQ); Reviewed: 05-May-2022, QC No. JBTW-22-61086; Revised: 10-May-2022, Manuscript No. JBTW-22-61086(R); Published: 20-May-2022, DOI: 10.35248/2322-3308.11.3.001
Copyright: © 2022 Du C, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.