Journal of Multiple Sclerosis
ISSN - 2376-0389NLM - 101654564
Mini Review - (2023) Volume 10, Issue 1
Introduction: The study of mechanisms that control gene expression without changing the underlying DNA sequence is known as epigenetics. DNA methylation is the most well-known. The cause of multiple sclerosis (MS), a condition with an unknown aetiology, has been hypothesised to involve environmental effects on individuals who already have a genetic susceptibility. DNA methylation may have a pathogenic effect at the point where genetic predisposition and environmental influences converge. A review of the literature on the impact that environmental risk factors for the onset of MS can have on several epigenetic pathways and the impact that these alterations have on the progression of the illness.
Conclusion: Understanding the role of epigenetic alterations in the aetiology of MS opens up a new area of study that could lead to the discovery of new biomarkers and treatment targets.
Multiple sclerosis • Epigenetics • Vitamin D • Smoking
Epigenetic regulatory mechanisms
Conrad Waddington, who lived from 1905 to 1975, coined the word "epigenetics" in the middle of the 20th century, but it wasn't until lately that it started to gain popularity as a new area of study. Today, it is regarded as a promising source of knowledge, particularly in the medical sector [1].
The study of mechanisms that control gene expression without altering the deoxyribonucleic acid (DNA) sequence is known as epigenetics. This field shows how phenotypic development is influenced by both genetic and environmental factors. During fetal development, epigenetic alterations are crucial for cellular and tissue differentiation because they alter how some genes are activated in response to environmental factors. These modifications take place as adults as well. Epigenetic alterations occur in human cells over the course of their existence. In reality, depending on the environmental elements they are exposed to, such as cigarettes, nutrition, or exercise, identical twins (with the same genetic load) accumulate different epigenetic patterns [2]. This translates into discernible variations in both twins' phenotypes, which show up as various susceptibilities to illness or disease outcomes.
The three main epigenetic mechanisms are non-coding RNA, DNA methylation, and histone modification. The most well-known mechanism is DNA methylation, and numerous research have examined how it may contribute to the emergence of illnesses. Our research is concentrated on this mechanism [3, 4].
DNA methylation
Cytosine residues in the RNA nucleotide chain are added methyl groups during the process of DNA methylation. Cytosine-guanine dinucleotides (CpG), which build up in the genome to create CpG islands, are the sites where this binding takes place. In gene promoter regions and other regulatory areas, these islands are very prevalent. DNA methyltransferases (DNMT), which catalyze the transfer of a methyl group from S-adenosyl-lMethionine (SAM) to carbon 5 of cytosine, carry out methylation. This process can take one of two paths: either DNMT3a or DNMT3b catalyse the establishment of a de novo DNA methylation pattern or DNMT mediates the maintenance of a genomic methylation pattern across subsequent DNA replication cycles. Cytosine residues in the RNA nucleotide chain are added methyl groups during the process of DNA methylation [5].
Cytosine-guanine dinucleotides (CpG), which build up in the genome to create CpG islands, are the sites where this binding takes place. In gene promoter regions and other regulatory areas, these islands are very prevalent. DNA Methyltransferases (DNMT), which catalyse the transfer of a methyl group from S-adenosyl-l-methionine (SAM) to carbon 5 of cytosine, carry out methylation. This process can take one of two paths: either DNMT3a and DNMT3b catalyse the establishment of a de novo DNA methylation pattern or DNMT mediates the maintenance of a genomic methylation pattern across subsequent DNA replication cycles [6].
As previously mentioned, the methyl group donor is the SAM molecule, which loses its methyl group to create S-Adenosyl-lhomocysteine (SAH).
SAM is hydrolized to homocysteine and then remethylated to methionine with the help of the cofactor 5-methyltetrahydrofolate (5-MTHF). Methionine Adenosyltransferase (MAT) is ultimately responsible for converting methionine into a SAM molecule (Figure 1). The ratio of the SAM to SAH levels in blood will determine the likelihood of DNA methylation [7].
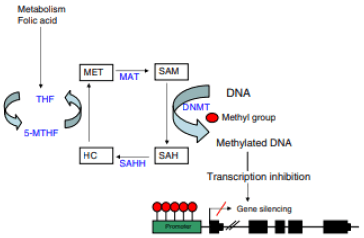
Figure 1: DNA methylation: schematic representation of the metabolic process involved. Homocysteine, methionine, methionine adenosyltransferase, S-adenosyl-l-homocysteine, homocysteine hydrolase, SAM, tetrahydrofolate, and 5-methyltetrahydrofolate are all abbreviations for DNA methyltransferases (DNMT).
The likelihood of DNA methylation increases with ratio. Therefore, we can conclude that homocysteine, methionine, other enzymes involved in this metabolic pathway, as well as chemicals like folic acid and vitamin B12, must all be properly metabolised for DNA methylation to occur.
Epigenetic pathways and smoking
Prenatal exposure to tobacco and higher methylation of the Brain Derived Neurotrophic Factor (BDNF) promoter, which encourages the differentiation and creation of new neurons, have been linked, according to a study that examined blood samples from adolescents whose mothers smoked during pregnancy. PCR testing were used to compare the DNA methylation patterns of smokers and non-smokers. They discovered alterations in DNA methylation in 438 genes in smokers and 25 genes in non-smokers. Studies of cancer patients have also discovered epigenetic alterations associated to smoking. Smokers showed hypermethylation of the CDKN2A, DAPK, and MGMT tumor suppressor genes in a research that included lung cancer patients. Similarly, smokers' cervical epithelial cells had hypermethylation of the CDKN2A gene, according to a study of cervical cancer in women between the ages of 15 and 19 [7].
DNA methylation's importance in clinical settings
In recent years, there has been an increase in interest in the relationship between human disease and changes in epigenetic mechanisms; evidence of such a correlation has been documented in a number of illnesses, particularly in the field of cancer [8, 9]. Colorectal Cancer (CRC) was the first kind of tumor to be linked to epigenetic regulatory mechanisms. When compared to cancer cells from healthy controls, cancer cells from individuals with CRC initially showed a reduction of overall methylation [10]. At the same time, it was discovered that the promoters of tumor suppressor genes were methylated, resulting in a reduction in the production of those genes [11]. These findings confirmed the link between disease development and increased methylation of tumor suppressor genes. However, it is yet unclear how altered DNA methylation contributes to the development of diseases in other medical specialties, such as neurological disorders. Epigenetic modifications that may play a role in the pathophysiology of Multiple Sclerosis (MS) have just been discovered, bringing up a fresh and intriguing area of study [11-13].
Young and middle-aged persons with significant neurological impairment are thought to be most commonly affected by MS. According to prevalence surveys, there are about 80 instances per 100,000 people in Spain. The Central Nervous System (CNS) is affected by the chronic disease MS, which causes inflammatory, demyelinating, and neurodegenerative lesions. MS is thought to have an autoimmune, likely multifactorial origin linked to a number of genetic and environmental susceptibility factors, while its aetiology is yet unknown. We can hypothesise that altered epigenetic regulation may play a role in MS given the complexity of the disease and the several aetiological processes (genetic and environmental) involved in MS pathogenesis.
A neurological condition known as multiple sclerosis places a heavy cost on healthcare systems, society, and families. Despite recent developments in our understanding of MS, the precise aetiopathogenic mechanisms underlying the illness remain unknown, and no effective cure has yet to be discovered.
Environmental variables can affect a person's gene expression through epigenetic modifications like DNA methylation. A fascinating area of medical research is epigenetics, particularly when it comes to illnesses like MS where environmental risk factors may be involved.
Despite the small number of published studies involving MS patients, the findings support more research in this field. The modulation of DNA methylation in candidate genes, which is essential for the emergence of MS, is implicated in the development of autoimmune disorders. Even though there is growing evidence of this link, more research needs be done using bigger patient and control populations.
Understanding the pathogenic mechanisms of MS will help us identify possible biomarkers and new therapy targets by elucidating the epigenetic modifications involved in the disease's a etiology.
Citation: Hardy J, Brown M. Neurology Epigenetic Changes: DNA Methylation in Multiple Sclerosis. J Mult Scler 2023, 10(1), 483
Received: 07-Jan-2023, Manuscript No. JMSO-22-96457; Editor assigned: 10-Jan-2023, Pre QC No. JMSO-22-96457 (PQ); Reviewed: 24-Jan-2023, QC No. JMSO-22-96457 (Q); Revised: 26-Jan-2023, Manuscript No. JMSO-22-96457 (R); Published: 29-Jan-2023, DOI: 10.35248/2376-0389.23.10.1.483
Copyright: ©2023 Hardy J. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.