Research - (2019) Volume 8, Issue 7
The Antiretroviral Therapy (ART) has reduced the mortality and morbidity of Human Immunodeficiency Virus 1 (HIV 1) in humans. The adverse effect of anti-retroviral therapy is associated with the adherence difficulty, substancial toxicity and abnormal cost. In the process of identifying a novel inhibitor to treat HIV, CCR5 (chemokine receptor 5) remains a novel target. Though there is an FDA approved inhibitor and crystal structure for CCR5, the resolution and interactions lacks explanation with respect to the site of action. Since the coverage and resolution of CCR5 was poor, Comparative modeling was performed to identify a homologues structure with complete coverage. In this study, we focus on identifying a lead to inhibit CCR5 by the process of virtual screening from various libraries of small molecules. Then the associated interactions of CCR5 and the lead molecule for were analyzed. Future works in the current research will be extended to perform meticulous studies on identifying the potency of interactions in a simulated environment, identifying the structure, property and activity relationships between the conformers of lead molecules.
ART, HIV 1, FDA, CCR5, Modeling, Virtual screening, Lead
Chemokine receptors and chemokines play a vital role in the trafficking of the populations of leukocyte across the body, and are involved in the development of a large variety of human diseases [1]. CCR5 is the main coreceptor utilized by the macrophage of the (M)-tropic strains of Human Immunodeficiency Virus type 1 (HIV-1) and HIV-2. CCR5 is an essential target in the process of HIV pathogenesis [2]. The antagonists of CCR5 include various inflammatory chemokines like RANTES, MCP-2, MIP- 1 alpha and MIP-1 beta. MCP-3 is a natural antagonist of the receptor. CCR5 is mainly expressed in macrophages, memory T-cells and immature dendritic cells [3]. CCR5 is upregulated by proinflammatory cytokines. It is coupled to the Gi class of heterotrimeric G-proteins, and inhibits cAMP production, stimulates Ca2+ release, and activates PI3-kinase and MAP kinases, as well as other tyrosine kinase cascades. A mutant allele of CCR5, CCR5 delta 32 is frequent in populations of European origin, and encodes a nonfunctional truncated protein that is not transported to the cell surface [4]. Homozygotes for the delta 32 allele exhibit a strong, although incomplete, resistance to HIV infection, whereas heterozygotes display delayed progression to acquired immunodeficiency syndrome (AIDS). Many other alleles, affecting the primary structure of CCR5 or its promoter have been described, some of which lead to nonfunctional receptors or otherwise influence AIDS progression. CCR5 is considered as a drug target in the field of HIV CCR5 inhibitors are a relatively new class of antiretroviral inhibitors used in the treatment of Human Immunodeficiency Virus (HIV). The inhibitors are designed to prevent the infection of HIV in the CD4 T-cells by blocking the receptor CCR5 [5]. When the receptor CCR5 is unavailable, the variant of the virus that is common in earlier HIV infection is called as the ‘R5-tropic’ and it cannot be engaged with a CD4 T-cell.
Galaxy Seok lab
The package of GALAXY server consists of various programs like (i) GalaxyTBM (the template-based modeling program), GalaxyLoop (the loop/terminus modeling program), GalaxyRefine (the model refinement program), GalaxyGemini (the homooligomer prediction program), and the Galaxy Dock (proteinligand docking program). These programs were tested with the critical assessment of techniques for Protein Structure Prediction. Developments of these programs were based on the global optimization of modeling with energy functions. The free energy functions of GALAXY were carefully designed by combining the principles of physical chemistry with the information of sequence and structure of protein. Confirmation search methods were based on the concept of annealing in space and the closure of triaxial loop [6]. In this work GalaxyTBM was used to perform the homology modeling of CCR5 with its template structure.
Molprobity
Molprobity server provides the validation of the molecular models of macromolecules [7]. The mirror site of MolProbity is available at http://molprobity.manchester.ac.uk. The open-source code is available at GitHub server. Two distinct data sets of distance criteria in heavy-atom-to-hydrogens accompanies the van der Waals radii were added in molprobity to improve accuracy. New validations were added to molprobity in Ramachandran plot and rotamer criteria. In this work, molprobity was used to validate the modeled structure of CCR5. In this work molprobity was used to analyze the quality of the modeled protein.
MTi open screen
MTi Open screening facilitates the identification of new bioactive compounds with an innovation in algorithm to improve the effectiveness of the processes of drug discovery. MTi Open Screen is dedicated to the process of virtual screening of small molecules. MTi Open screen performs the automated virtual screening with AutoDock Vina. MTiOpenScreen provides the valuable collections of the drug-like libraries of chemical containing 1,50,000 compounds in PubChem for screening [8]. In this work MTI open screen was used to perform virtual screening.
Swiss dock
Swiss Dock is a web server for docking small molecules on target proteins. It is based on the algorithm of EADock implemented in the DSS engine and gets combined with the Ajax scripts for curating the common problems for preparing the input files of the target protein and the ligand. The web site provides an access to a database of manually curated complexes [9]. In this work Swiss Dock was used to perform docking.
Ligplot+
LigPlot+ is the successor of the original LIGPLOT program for the automatic generation of 2D interactions between the ligand and the protein. It runs from a java interface and allows the on-screen editing of the plots via click-and-drag operations in mouse [10]. In this work LigPlot+ was used to perform the interaction analysis of hydrogen bonding, vander wall and coloumb interactions.
Homology modeling of CCR5 in galaxy Seok lab server
The crystal structure of CCR5 (PDB Id 5 UIW A) was used as a template to construct the homology model of CCR5. The modeled structure of CCR5 is given in Figure 1.
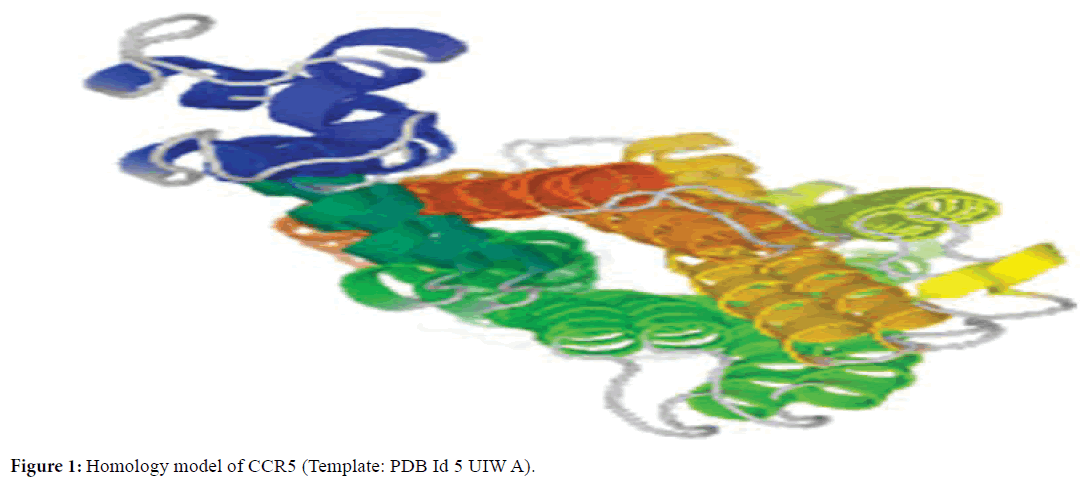
Figure 1. Homology model of CCR5 (Template: PDB Id 5 UIW A).
Structural validation of CCR5
Molprobity server was used to perform the structural validation of CCR5. The structural analysis of Ramachandhran Map in Molprobity is given in Figure 2. The analysis of Ramachandhran Plot confirms that there were no residues present in the out layer and more than 98% of residues were present in the favored and allowed regions.
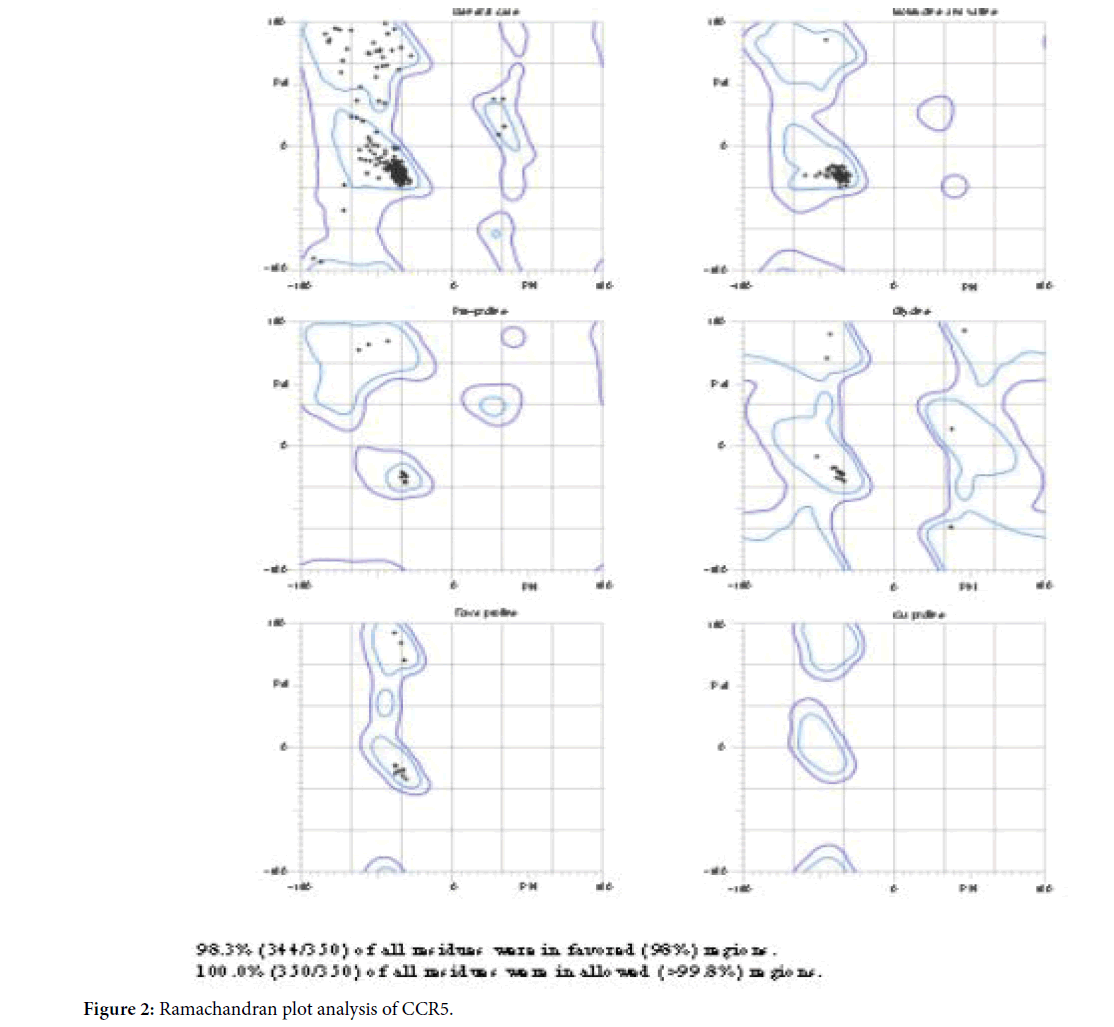
Figure 2. Ramachandran plot analysis of CCR5.
Virtual screening of CCR5 in MTI opens screen
In MTI open screen, virtual screening was performed for CCR5 on the basis of Lipinskis rule with the diverse drug library of 1,50,000 compounds. The process of virtual screening resulted in identifying 1500 compounds as initial leads. Then the best 10 compounds were tabulated in Table 1 on the basis of their binding free energy.
| Compound (ZINC database ID) | Energy (Kcal/mol) | nRot | HBA | HBD | LogP | MW | TPSA |
|---|---|---|---|---|---|---|---|
| 24324394 | -8.5 | 5 | 4 | 1 | 4.86 | 442.5725 | 74.71 |
| 17505192 | -8.4 | 4 | 6 | 1 | 4.09 | 413.859 | 72.18 |
| 24280440 | -8.3 | 2 | 7 | 0 | 2.63 | 387.3881 | 83.72 |
| 24286776 | -8.2 | 4 | 6 | 0 | 4.76 | 479.4969 | 53.74 |
| 47195657 | -8.2 | 4 | 6 | 2 | 4.05 | 446.3975 | 90.44 |
| 863204 | -8.2 | 2 | 8 | 1 | 2.35 | 393.7833 | 97.19 |
| 17449993 | -8.1 | 1 | 6 | 2 | 2.3 | 335.3566 | 95.94 |
| 14746093 | -8 | 5 | 8 | 3 | 2.44 | 478.8085 | 113.06 |
| 24840422 | -8 | 5 | 9 | 3 | 2.81 | 452.5294 | 142.78 |
| 1.03E+08 | -8 | 5 | 5 | 1 | 4.48 | 365.4305 | 55.11 |
Table 1: Best leads of CCR5 from virtual screening.
Among the leads, the compound 24324394 (ZINC database Id) was more stable and it is subjected to docking in Swiss dock.
Docking of CCR5 and compound 24324394 (ZINC database Id) in Swiss dock
In Swiss dock, CCR5 was docked with the compound 24324394 (ZINC database Id) and around 10 poses were obtained. Among the 10 poses, the best pose were selected and given in Figure 3.
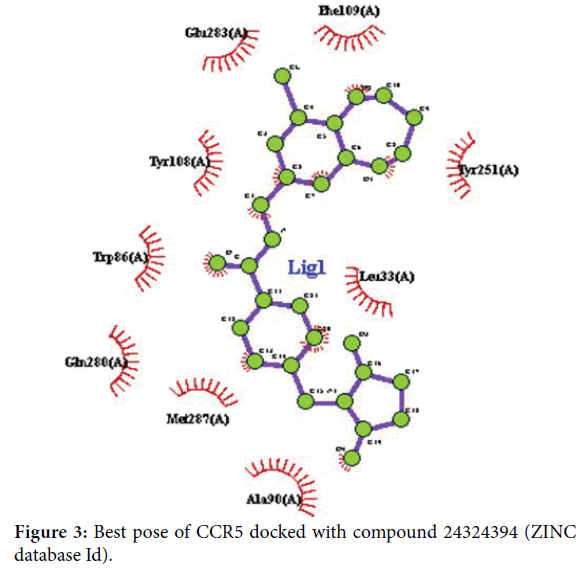
Figure 3. Best pose of CCR5 docked with compound 24324394 (ZINC database Id).
Interaction analysis CCR5 docked with compound 24324394 (ZINC database Id) in Ligplot
VanderWalls and Coloumb interactions were analyzed within 5Å and the interacting residues were identified. The list of interacting residues was given in Figure 4.
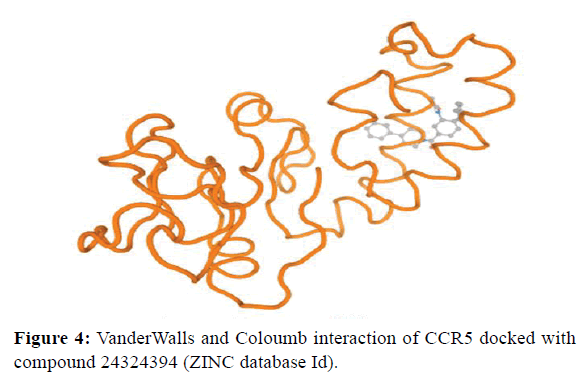
Figure 4. VanderWalls and Coloumb interaction of CCR5 docked with compound 24324394 (ZINC database Id).
Designing a potential inhibitor for CCR5 to compete the spread of viral load in HIV is a challenging task to execute. In this manuscript the challenge was addressed by modeling the complete structure of CCR5 with the template of the existing crystal structure. Then the quality of modeled structure is analysed by Ramachandran Plot. Then the modeled structure was subjected to virtual screening to obtain leads. Then a specific lead molecule was selected and docked with CCR5. Finally the interactions of small molecule and CCR5 were analyzed. This study provides a basic understanding of the process of rational drug design to identify leads for designing inhibitors CCR5. Future works in the current research will be extended to perform meticulous studies on identifying the potency of interactions in a simulated environment, identifying the structure, property and activity relationships between the conformers of lead molecules prior to experimental studies in animals and humans.
Received: 12-Sep-2019
Copyright: © 2019 Harishchander A, et al. This is an open access paper distributed under the Creative Commons Attribution License. Journal of Biology and Today's World is published by Lexis Publisher.